How “Quality Control” Functions in Ribosome Assembly
19 November 2013
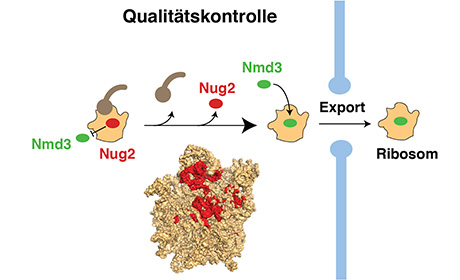
Image: Ed Hurt
Like automobiles, ribosomes must pass through “quality control” after their many components are assembled. Only then can they be taken to their destination and proceed with their task – making proteins in the cell. A team of scientists led by Prof. Dr. Ed Hurt at Heidelberg University Biochemistry Center investigated an important component in this control process. The researchers demonstrated that a specific protein, which occurs in all cells from yeast to humans, works like a molecular control switch and prevents incomplete ribosomes from leaving the “assembly hall”. The team’s findings have been published in “Nature“.
Ribosomes are macromolecular complexes consisting of a large number of parts comprising ribonucleic acids and ribosomal proteins arranged in a specific three-dimensional structure. Correct ribosome assembly is essential for the survival of all cells. The assembly process for these many components is subject to strict rules, similar to an automobile assembly line. Before the finished car leaves the assembly hall, it is tested for full functionality and then sent to its destination. According to Prof. Hurt, there has been little research thus far on how this “quality check” takes place in ribosome assembly. The Heidelberg scientists have now identified the function of a protein from the energy-consuming GTPase family in this process.
Only correctly assembled ribosomes make it from the “assembly hall” in the cell nucleus to the cytoplasm, where they set about their real task of synthesising proteins. For export to the cytoplasm, nascent ribosomes must pass through a quality checkpoint for binding to export factor Nmd3. Before binding, however, a signal has to be given that the assembly of the ribosome is correct and complete. Until then, a type of “control officer” occupies the site earmarked for Nmd3. The Heidelberg team has now demonstrated that this function is performed by a member of the GTPase protein family known as Nug2. Only when a specific enzyme swings into action to remodel the assembled ribosome for export to the cytoplasm, thereby consuming the energy stored by the GTPase, does the Nug2 “control officer” vacate its place and make way for export factor Nmd3. This is how the export of incomplete ribosomes is prevented.
Now that Prof. Hurt’s team has been successful in identifying the role played by control agent Nug2 in ribosome assembly, the researchers hope to gain even further insight into the functions of related proteins. These so-called nucleostemins are found in higher eukaryotes, including those of humans. The Heidelberg scientists plan to decipher the molecular mechanisms that various enzymes from the nucleostemin family use to combine the two fundamental cellular processes of ribosome formation and cell proliferation. Scientists at the University of Edinburgh (UK) also cooperated in the current research.
Original publication
Y. Matsuo, S. Granneman, M. Thoms, R.-G. Manikas, D. Tollervey and E. Hurt: Coupled GTPase and remodeling ATPase activities form a checkpoint for ribosome export, Nature (17 November 2013), doi:10.1038/nature12731

