Current Research
While our focus in the first period at EMBL (1999-2002) was mainly on early pattering, the second period (2003-2007) is characterized by key findings in optic vesicle (Rembold et al., 2006) and optic cup (see below) morphogenesis. Our gain of function experiments had indicated that the homeodomain containing transcription factor Six3 is sufficient for ectopic eye formation (Loosli et al., 1999). In a loss of function analysis we showed that its activity is also necessary for the formation of the entire forebrain and eyes (Carl et al., 2002). Interestingly Six3 also plays subsequent roles related to the proximodistal patterning of the eye field. Our analysis indicates that, depending on the interactor, Six3 determines different fates along the proximo-distal axis (Tessmar et al., 2002) and, by direct interaction with the replication initiation complex, positively acts on proliferation (Del Bene et al., 2004).
This interaction interestingly shows the direct interference of a transcription factor with the cell cycle and conversely allows the cell cycle to modulate the transcriptional potential of a key player in eye development. In the absence of a battery of Six3 transcriptional targets the experimental basis for this far-reaching possibility was however thin. We have further pursued this issue and have identified Six3 target genes by ChIP from FAC sorted Six3-positive cells out of the embryonic context. In collaboration with colleagues form the Genome Institute of Singapore (Yijun Ruan, Chia-Lin Wei, GIS) we analyzed Six3 targets in a genome wide survey and (among many others) could show the direct binding of Six3 to its own upstream regions.
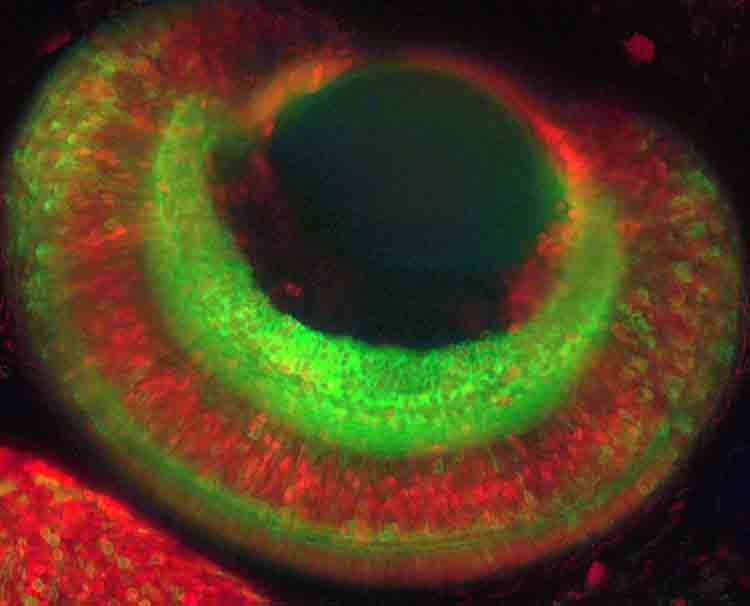 |
| Fig. 1: Overview of the eye of a double transgenic medaka fish at 4 days post fertilization. Green cells are expressing a membrane localized YFP under the control of the ath5 promoter, labeling the retinal ganglion cell layer. Ciliary marginal zone, müller glia and photoreceptors are labelled in red by H2B-RFP under the control of the rx2-promoter. The image is a maximum projection of a confocal stack recorded with the Leica SP5 and a 20x glycerol objective. |
Eye Morphogenesis
Optic vesicle evagination, the subsequent step in eye development, is impaired in the eyeless (el, medaka) and chokh (chk, zebrafish) mutants (Loosli et al., 2003; Winkler et al., 2000). The gene affected is another homeodomain containing transcription factor, Rx3 (Loosli et al., 2003; Loosli et al., 2001). Our analysis facilitated the generation of transgenic lines (in medaka and zebrafish) that express GFP specifically in the eye field of wild type and mutant embryos. These lines allowed following optic vesicle formation in vivo by 4D confocal analysis (Huisken et al., 2004; Rembold et al., 2006), one of our milestones in the past four years. In these studies we unraveled that organ morphogenesis crucially depends of the migratory activity of cells that, in contrast to the text-book view, migrate as individual cells in the context of eye formation (Rembold et al., 2006).
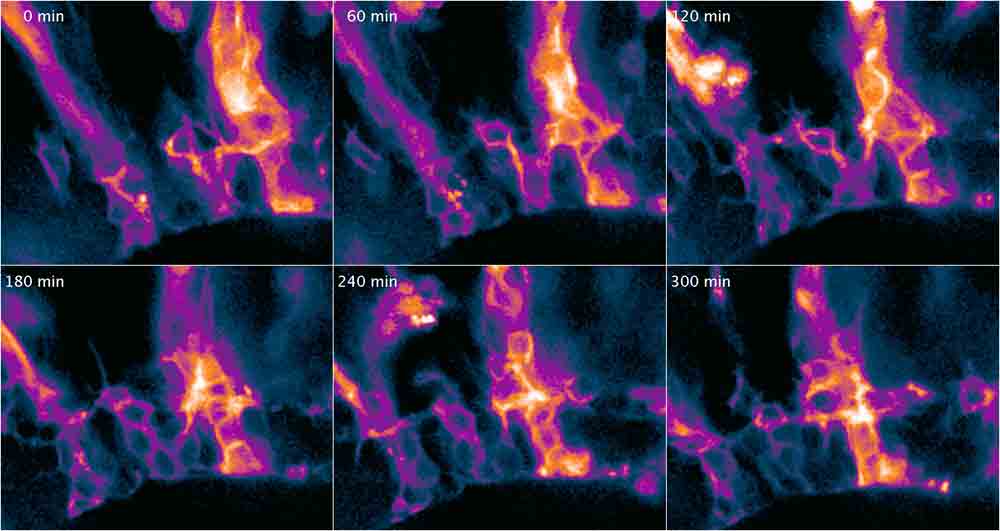 |
| Fig. 2: Time-lapse montage of lyn-tandem tomato expressing cells in the retina under the control of the vsx3 promoter. Columns of precursor-cells develop from a vertical to a horizontal network forming the inner plexiforme layer in the retina. Cells were imaged live and in vivo using a Leica SP5 and a 20x oil immersion objective. |
Eye Evolution
The work on Rx genes had led us to also investigate their role in basal Prototomia. We found that in the larva of the basic Lochtrophozoan Platynereis, Rx is expressed in ciliary photoreceptors, as in the eyes of Deuterostomia. These photoreceptors that are not part of the eye, are found in a complex network within the conserved axonal scaffold. This and other data led us to propose that the body plan of Urbilateria was quite elaborated (Arendt et al., 2001).
It contained already a visual complex at the anterior end of a conserved axonal scaffold that served as the evolutionary substratum from which the eyes of Proto- and Deuterostomia emerged (Arendt et al., 2004).
Medaka Genomics
To further develop medaka into a competitive model system in an efficient and economical way, it was of key importance to strengthen our contacts to the Japanese medaka community. We have been successfully collaborating with the Kondoh differentiation project completed the first description of mutants from a joint mutagenesis screen published in the “Medaka issue” of Mechanisms of Development (June/July 2004).
This and other activities catalyzed the initiation of a genome project that covers all aspects of medaka genomics. The medaka genome has now been released and should be published (in Nature) by the time of the review. With the expected publication of the zebrafish genome in the near future, almost 150 million years of evolution will be experimentally amenable in these fully developed model systems, putting fish into a unique position among the vertebrates.
We have started to establish ourselves in the field of bioinformatics to complement and direct bench work. At the moment we focus on mining the genomic information available in the context of gene and genome evolution (Martinez-Morales et al., 2007) and transcriptional networks related to eye development and evolution (Ettwiller et al., 2005; Henrich et al., 2003; Henrich et al., 2004; Henrich et al., 2005) in order to establish a virtuous cycle between bench and computer to direct future experimental work in a more system wide context.
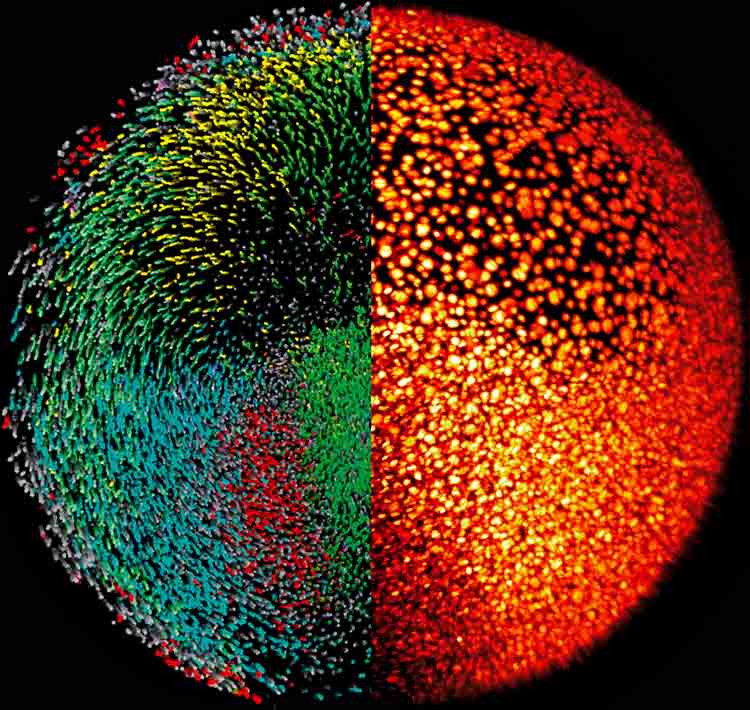 |
| Fig. 3: Reconstruction of the zebrafish digital embryo. The right half of the embryo is shown as the microscopy data, where nuclei are labeled using H2B-GFP (yellow/red LUT). The left half shows the segmented and rendered data with colors encoding movement directions of the individual cells (Keller, et al. 2008) |

