Current Research
The lab is currently addressing the underlying logic of cis-regulatory grammars governing precise spacio-temporal transcription during the establishment of neuronal diversity in vertebrates. The strategy consists of testing sequences that are likely to be involved in gene regulation using the in vivo enhancer assay that we developed and successfully tested. These data will dramatically expand the catalogue of fish enhancer conserved with mammals that have been characterised in vivo.
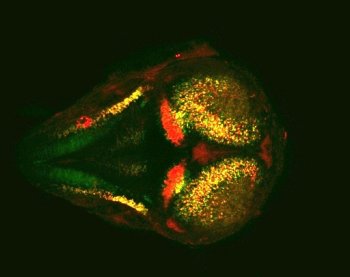 |
| Fig. 1: Enhancers of similar motif composition have similar activities. Double transgenic medaka embryo (dorsal view, stage 32) with Cherry reporter gene under the control of one enhancer and GFP reprter gene under the control of a similar enhancer. |
Effectively, the result of that analysis is the precise description of the outcomes (exemplified by the reporter gene expression in the medaka embryo) of many combinatorial regulatory elements hardcoded in these sequences. We therefore identify, for each neuronal structure studied, a number of possible combination of elements defining the necessary instruction for the activation of transcription in that structure. The integration of all the correct instructions is telling us:
- the key cis-regulatory elements and their relative arrangement within the sequences;
- to what extent these syntactical rules are flexible.
If these elements are necessary for the proper activation of transcription in a given structure, mutations affecting one or many of these element(s) would affect the enhancer activity of the regulatory sequence. We therefore perform site-directed mutagenesis in the elements that define the learned rules and test the modified sequences for their enhancer activities.
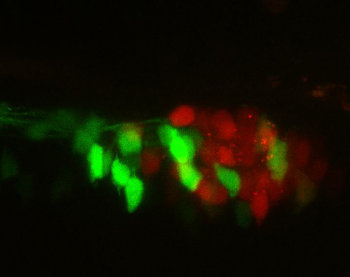 |
| Fig. 2: Slightly different motif composition in enhancers lead to activity in different cell type. |
Additionally, if these rules are necessary and sufficient, other regions satisfying these sets of rules would display enhancer activities similar to the initial validated sequences. We are also developing algorithms that search genomes for regions functionally similar to the validated enhancers. Sequences from various genomes can be screened for enhancer activity, opening this analysis to a more evolutionary perspective of cis-regulatory grammar.

