Current Research
Wnt signalling and brain asymmetry
Morphological and functional asymmetries in the central nervous system are a universal feature in the animal kingdom important for cognitive performance and social behaviour. The bilaterally formed habenular nuclei in the diencephalic epithalamus in vertebrates are part of a highly conserved conduction system and relays information from the telencephalon and diencephalon into the ventral midbrain. Proper function of this neural circuit is clearly important to man as alterations can have devastating consequences, such as depression or schizophrenia. Each habenula can be subdivided into lateral, medial and ventral neural cell populations, which send efferent axonal projections into different targets within the ventral midbrain, the interpeduncular nucleus (IPN) and raphe nuclei (Amo et al., 2010; Figure 1). Using lypophilic dyes and focal electroporation techniques to trace single efferent axonal projections of the habenula nuclei in the zebrafish we found that projections from these three habenula cell populations differ in their axonal morphology, branching and topology within their targets (Bianco et al., 2008).
Like in many other vertebrates the left and right habenula in zebrafish differ in the extend of neuropil and the expression of various marker genes. This asymmetric development is crucially dependent on precise regulation of Wnt/beta-catenin signalling, involving Axin1. Upregulation of this pathway by a mutation in the Axin1 gene in zebrafish, results in the loss of habenula asymmetry (Carl et al., 2007).
We are using molecular, genetic, transgenic and imaging techniques combined with next generation sequencing to understand about the mechanism how seemingly symmetric Wnt signalling (Beretta et al., 2011) is regulating asymmetric habenula development.
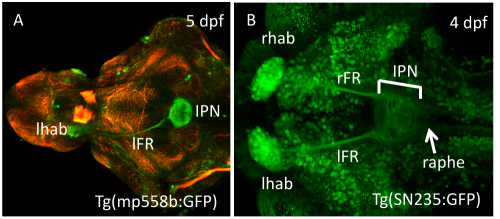 |
| Fig. 1: Transgenic resources allow in vivo studies Dorsal view with anterior to the left. Stage and name of transgenic lines are shown. (A) Dissected brain labeled with a-acetylated tubulin (red; picture taken by Kate Edwards). (B) Embryo showing GFP expression in all habenula cells, efferent projections and IPN (bar) and raphe (arrow; transgenic line generated by S. Ryu). dpf, days post fertilization; lFR, left fasciculus retroflexus; lhab, left habenula; IPN, interpeduncular nucleus; rFR, right fasciculus retroflexus; rhab, right habenula. |
Wnt signalling and axis formation
The emerging field of comparative developmental genetics studies distantly related, but morphologically similar species to identify conserved as well as species specific genetic and molecular mechanisms that underlie development and evolution in vertebrates. Concepts developed here are important because it will also be applicable to man - only if we can identify conserved genetic mechanisms we will be able to understand more about genetically derived diseases.
Medaka and zebrafish fulfill all requirements for a comparative approach: They are morphologically similar and evolutionary distantly related. Our aim is to functionally compare genes in these two teleosts that are required for early events important for posterior axis formation (Yokoi et al., 2007).
One widely used method to identify genes important for certain developmental processes in genetic model systems like fish is to mutate the genes so they no longer work and examine the consequences on embryonic development. In this process of ENU-mutagenesis point mutations are randomly introduced into the genome. In medaka, we have recently identified four fish lines with disturbed development of the posterior body axis (Figure 2). The fact that two of these show morphological alterations that have never been discovered so far in any vertebrate underscores the importance of using two distantly related species for developmental and evolutionary studies.
We have identified one of the genes mutated to be involved in the Wnt/beta-catenin signalling pathway. We are now combining a variety of genetic, transgenic and imaging techniques to elucidate and compare the function of this gene in both medaka and zebrafish.
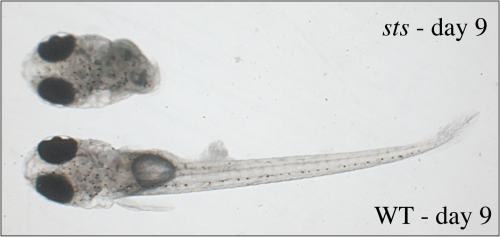 |
| Fig. 2: The sts mutant develops only a head while trunk and tail structures are absent The developmental stage is indicated; anterior is to the left. |

